Experiment
Purpose
By chemically modifying the template DNA with amino acids, we aim to modulate the oscillation period of the DNA-based system.
Methods & Result
About DNA
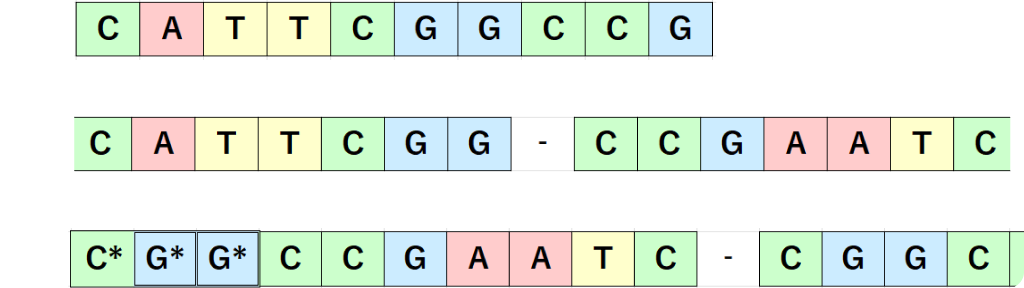
- Three types of DNA are used. Their base sequences are as follows.[1]
- A G strand modified with an amino group at the 3' end was also used for amino acid conjugation.
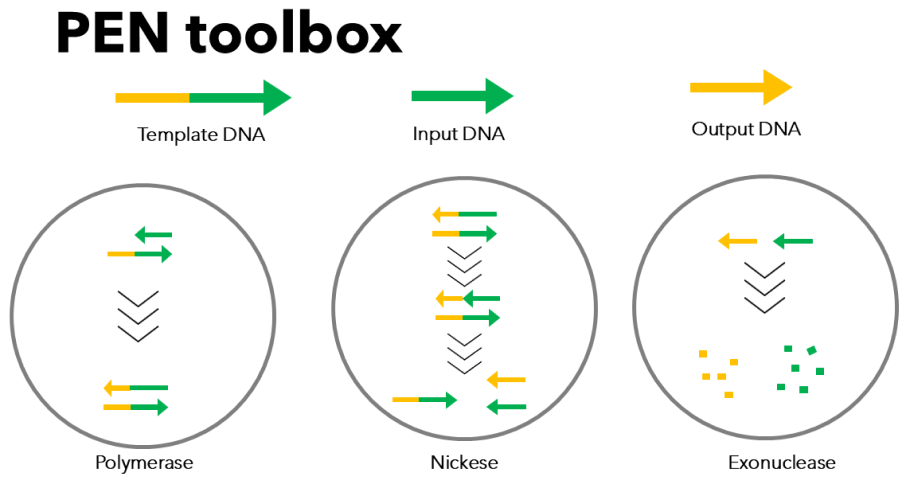
DNA PEN toolbox
- The nonlinear framework developed by Montagne et al. combines DNA-DNA hybridization reactions with enzymatic processes, enabling the construction of complex reaction network models analogous to gene regulatory networks. [2][3]
- In this system, a primer DNA hybridizes with a template DNA, is extended by a polymerase, cleaved by a nickase, and degraded by an exonuclease, thereby establishing a dynamic molecular system.
- Such a framework has been applied to the design of molecular oscillators and molecular automata.[4]
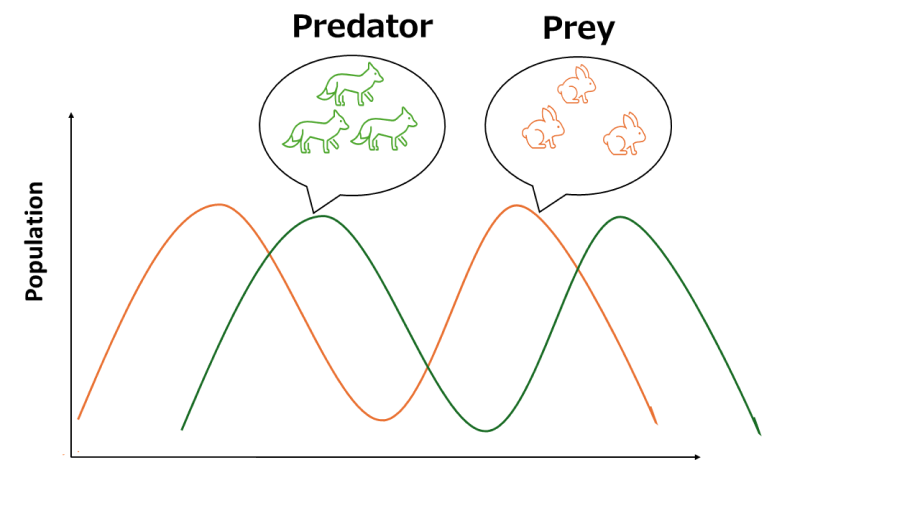
Predator Prey model
- A model representing the population dynamics between predators and prey in an ecosystem.
- As an example, the Lotka-Volterra equations can be cited.[1]
- The ratio between predators and prey transitions periodically over time.
- Using the PEN toolbox, this model is applied to DNA oscillation.
- In this framework, each DNA species has a specific role:
- P: Predator
- N: Prey
- G: Template
DNA-oscillation
- An oscillatory system modeled on the Lotka-Volterra equation.[1]
- Chemical oscillations are formed by constructing and combining Reactions 1-3 below.
Reaction 1 (Reaction indicating amplification of N)
- G represents template DNA, and N represents primer DNA. They bind to each other.
- Polymerase extends the sequence from the 3' end of N.
- Nickase cleaves the middle of the extended N sequence.
- Dissociation increases the number of Ns by one.
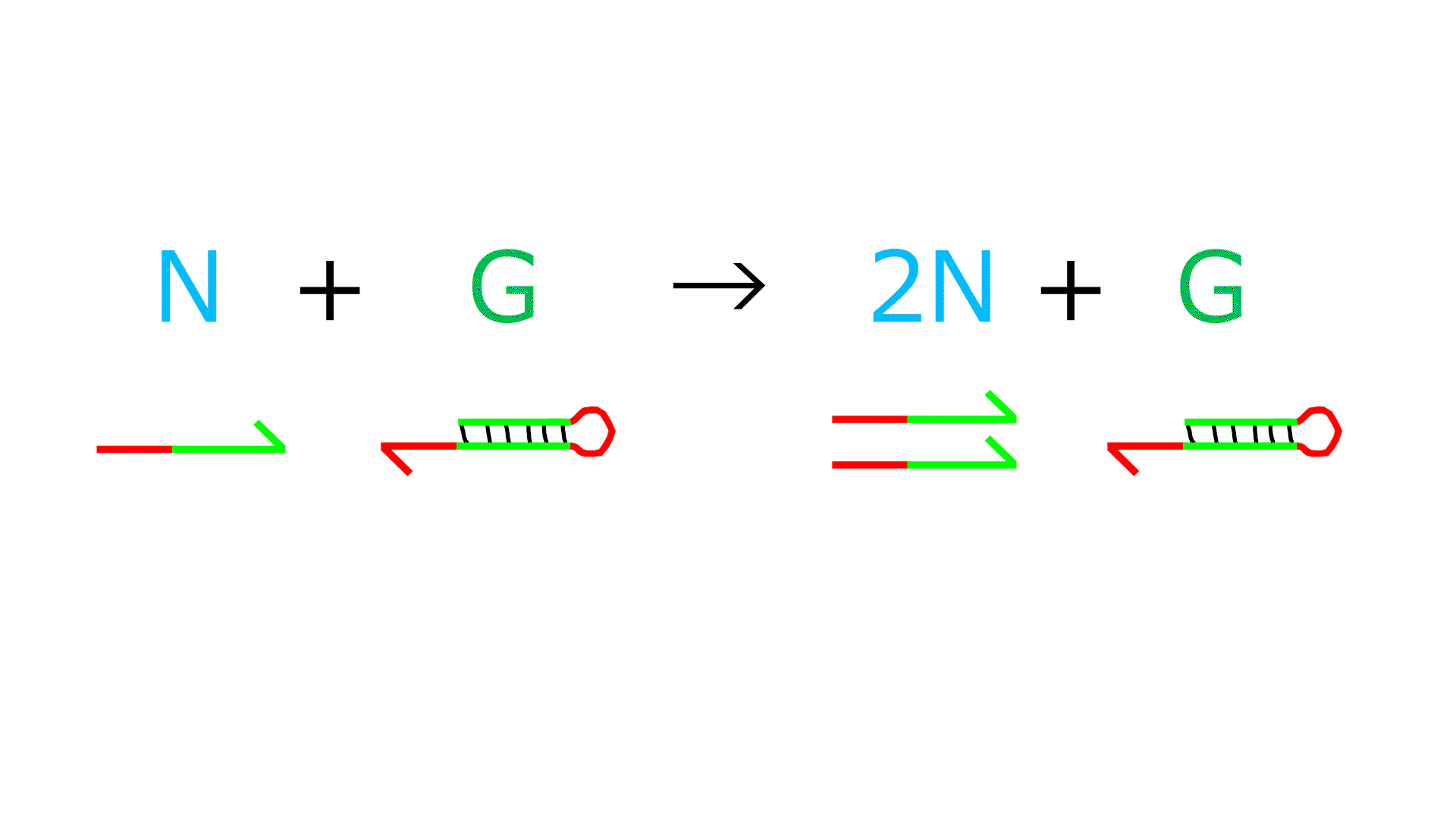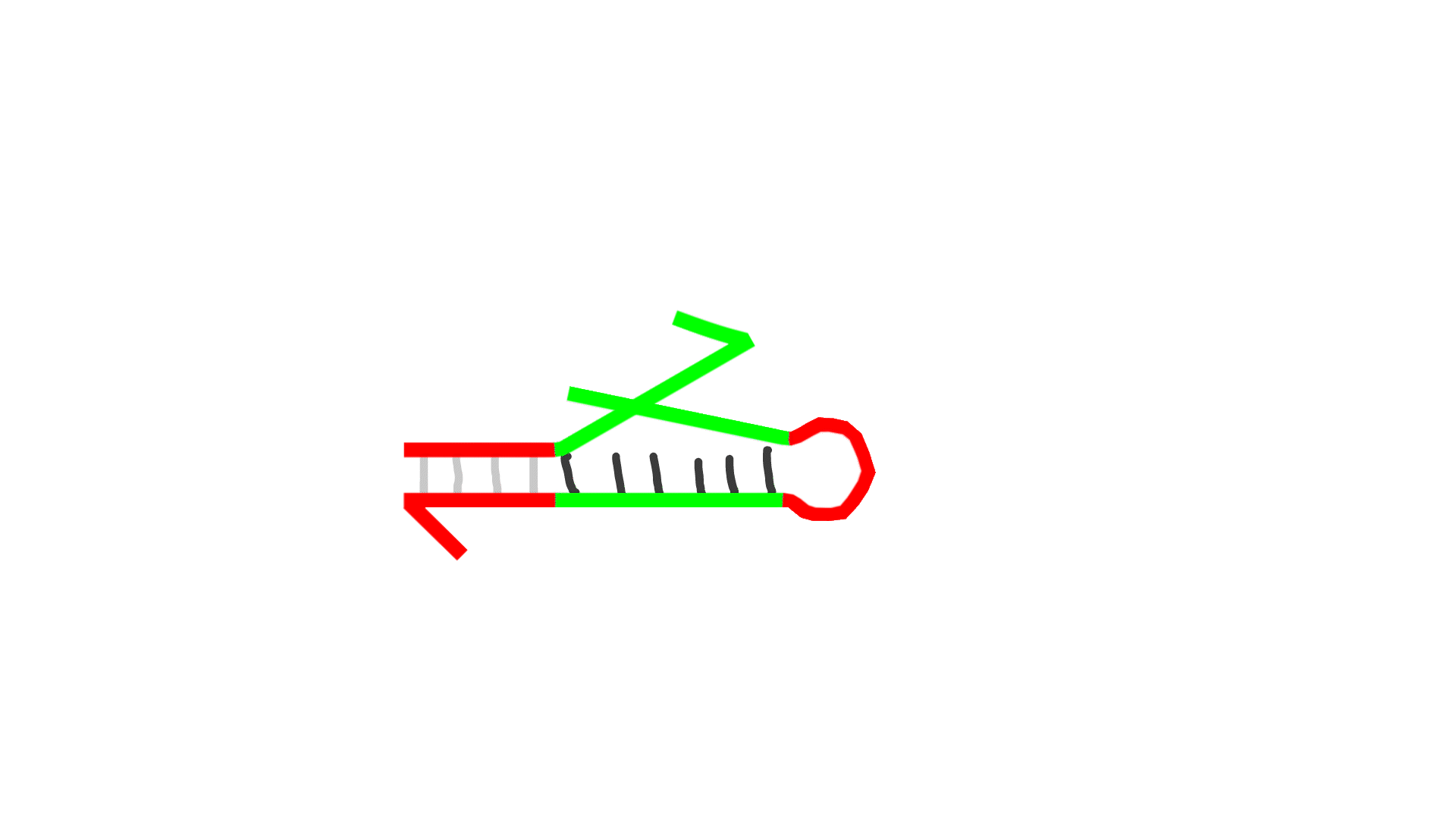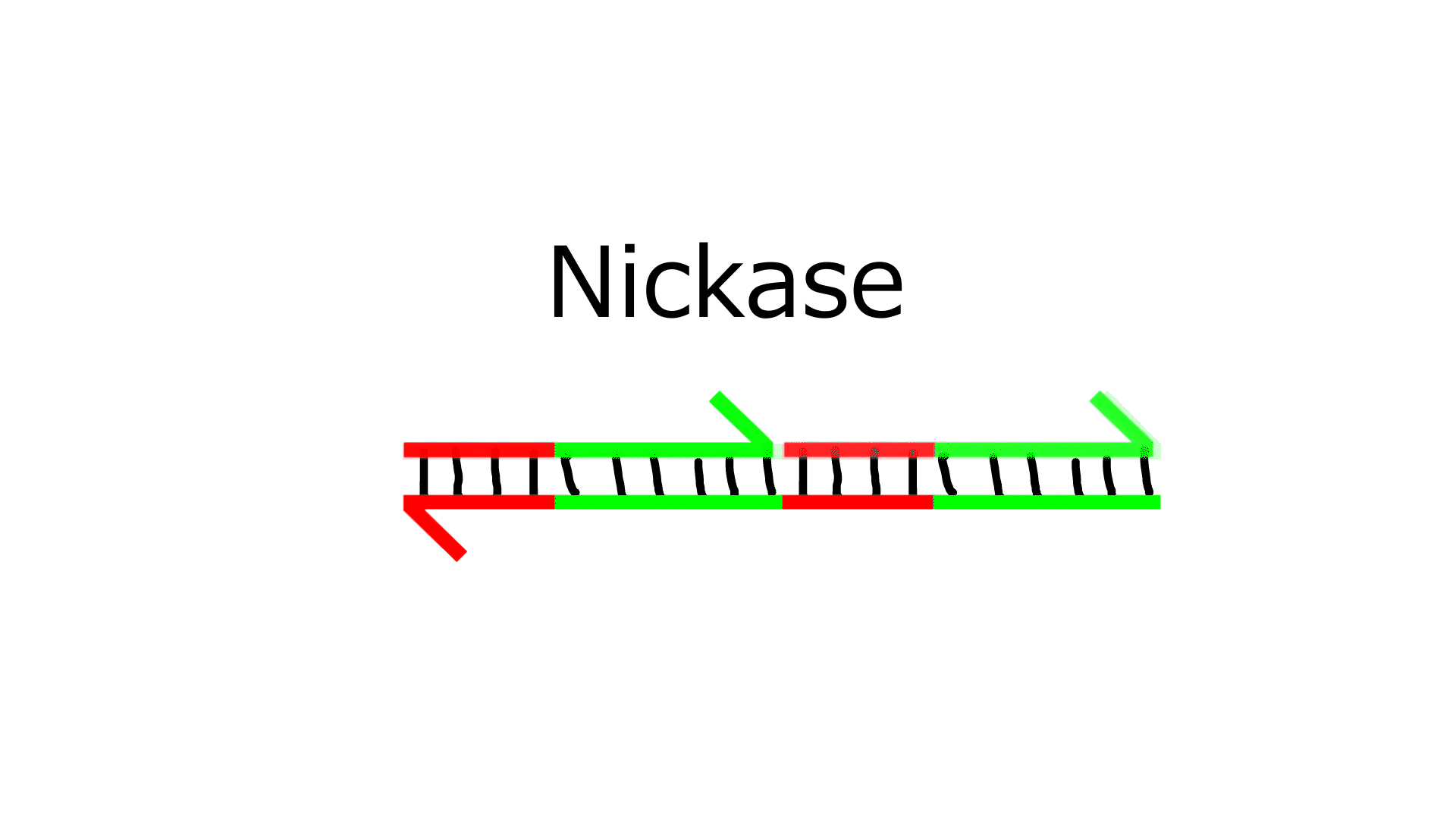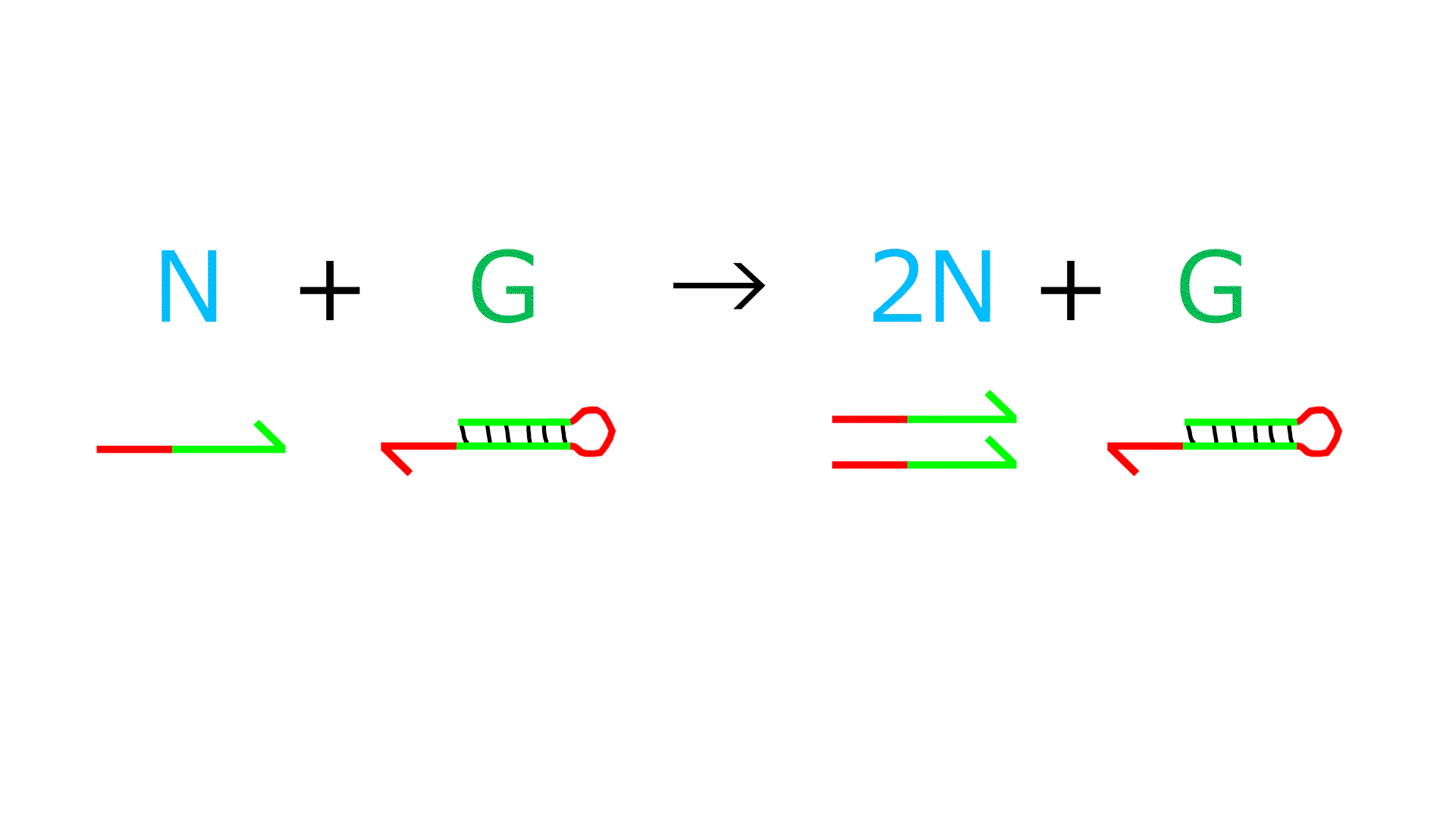
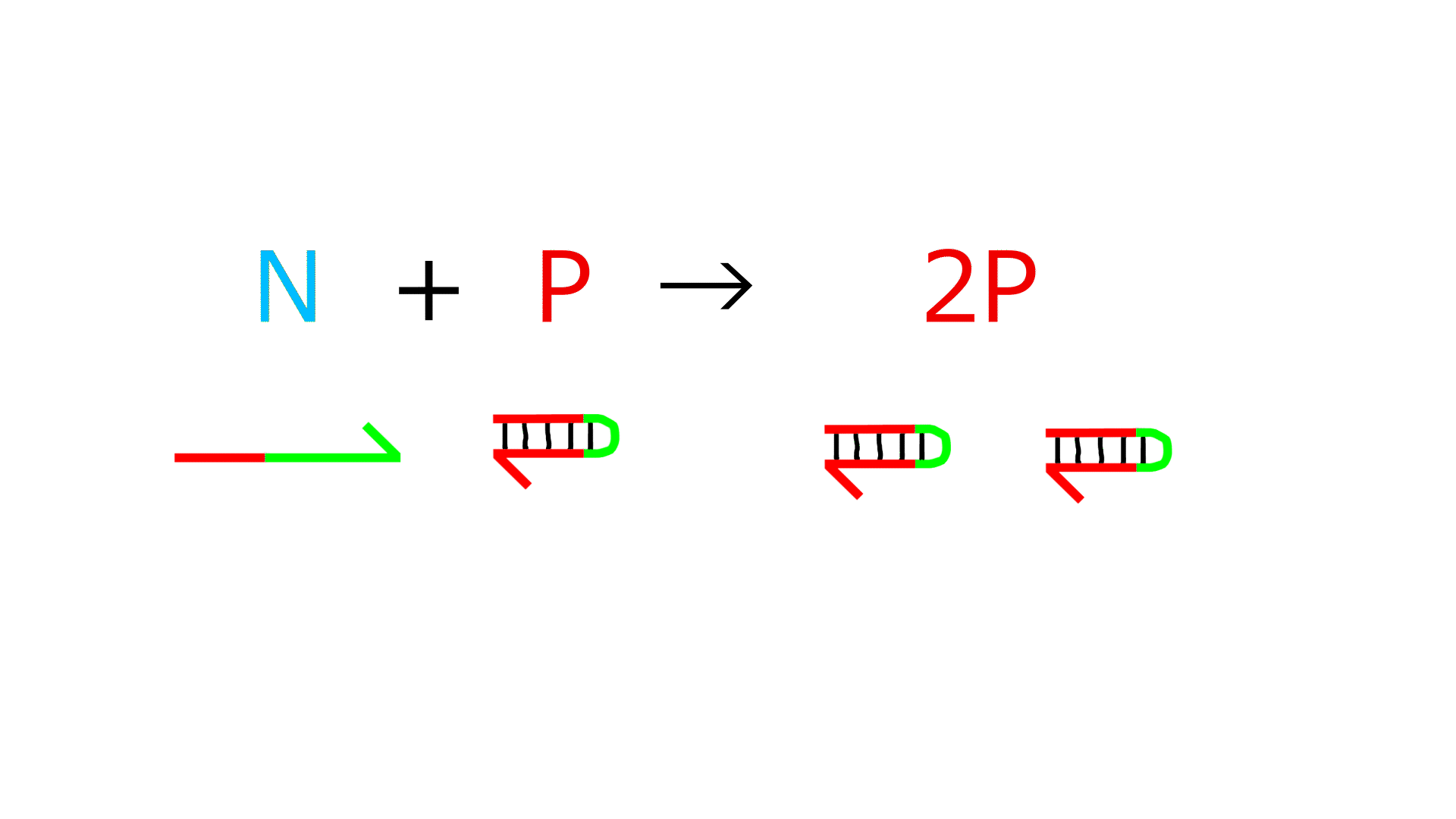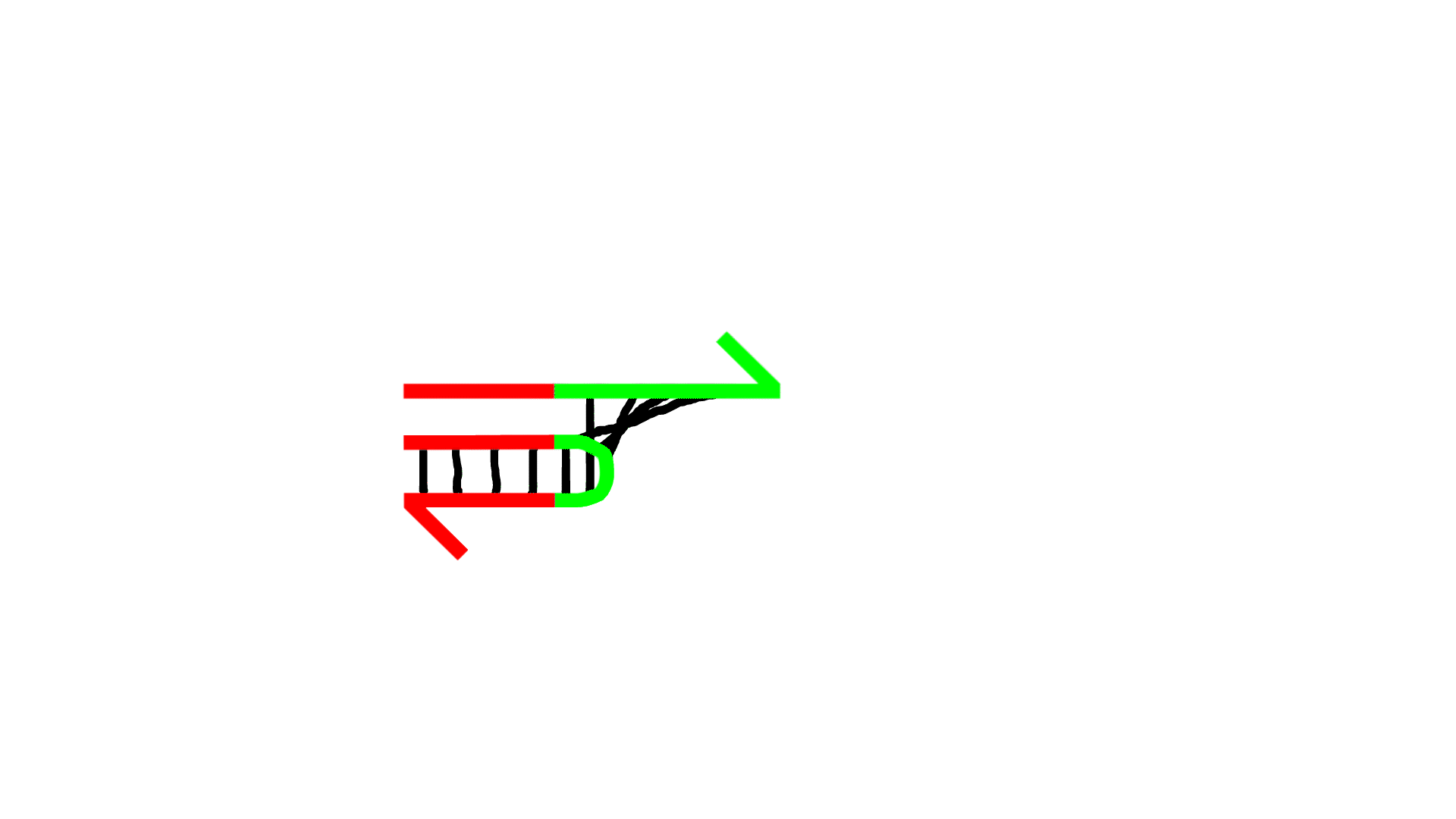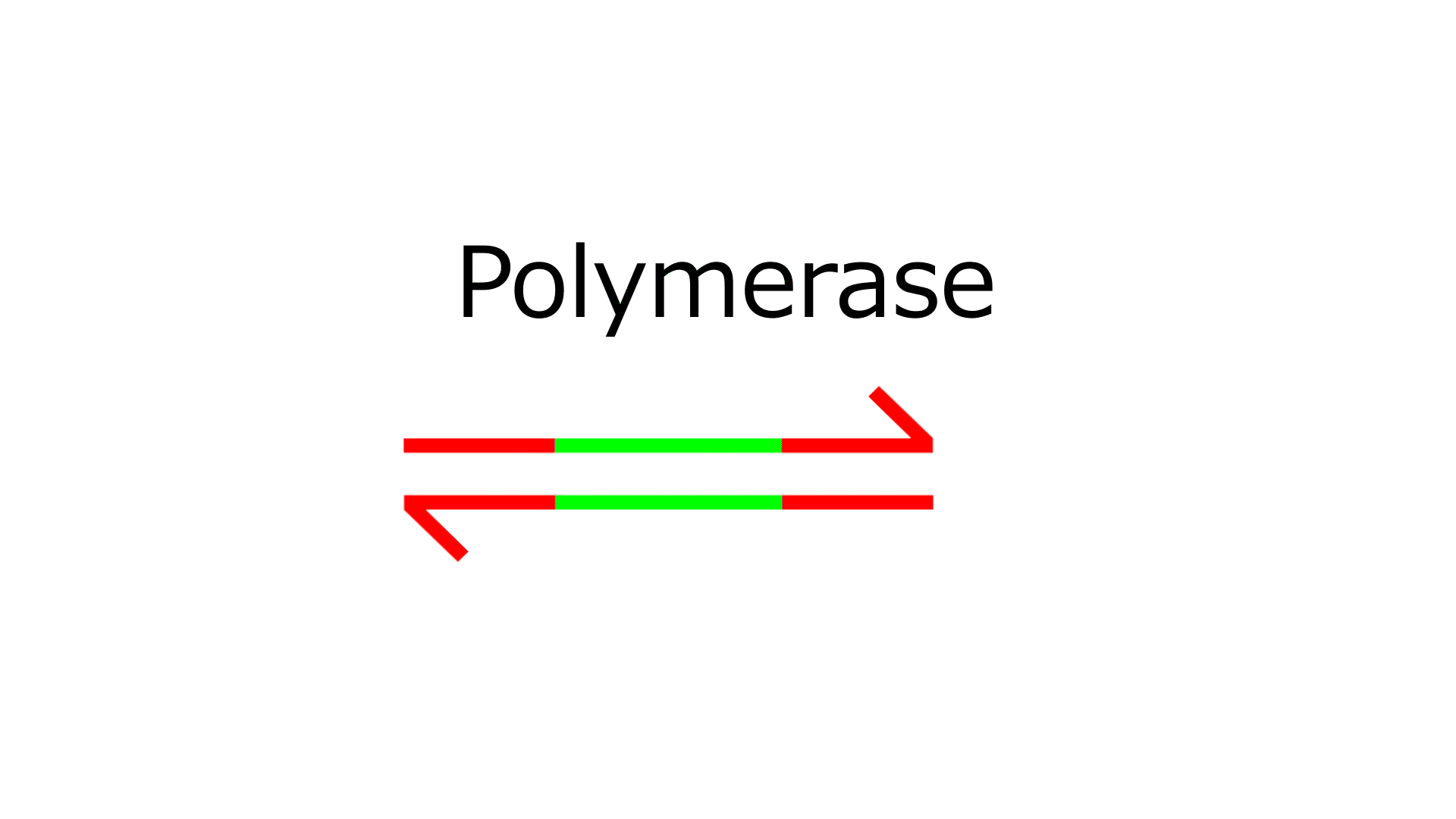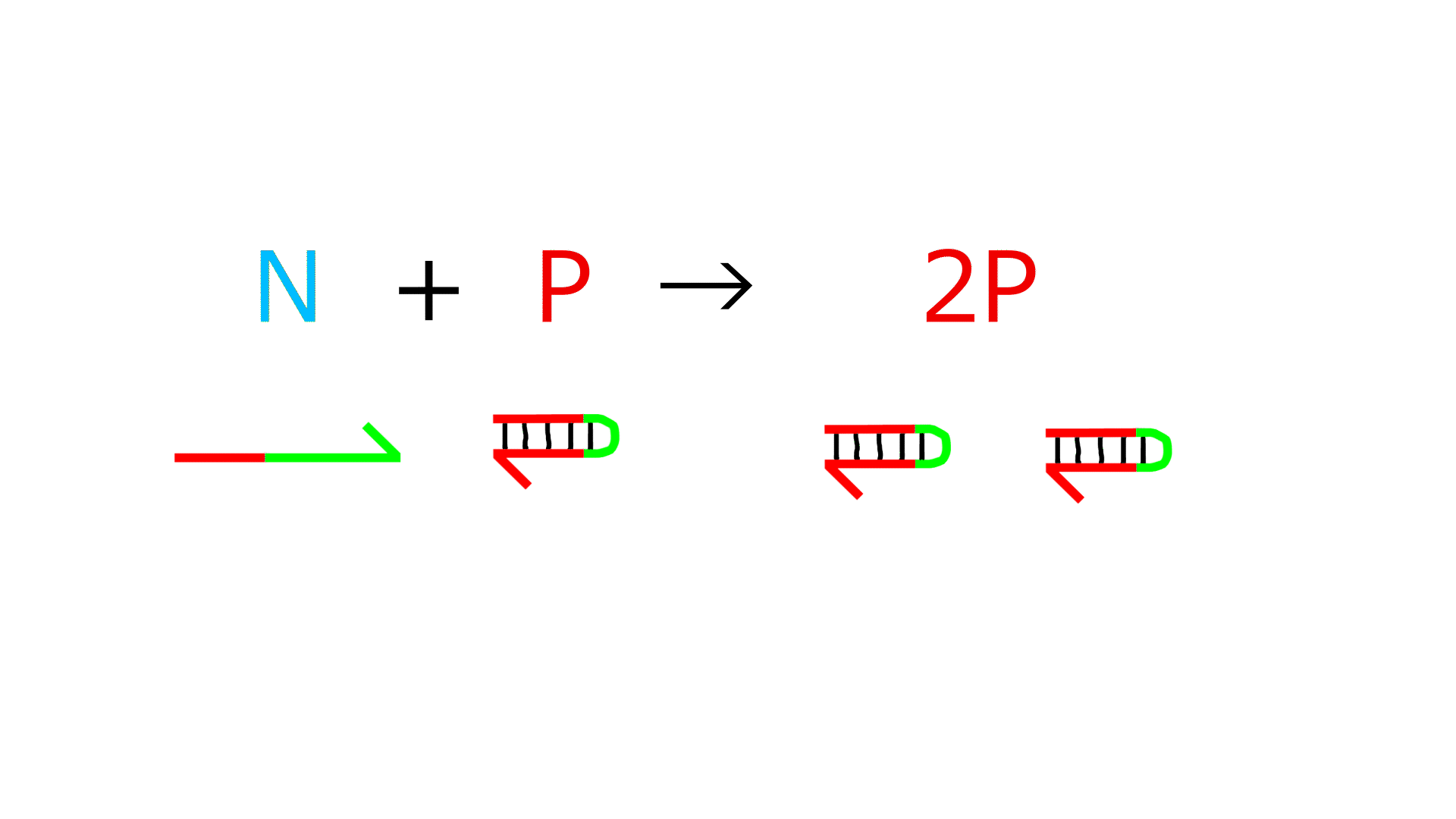
Reaction 2 (Reactions showing amplification of P)
- The P sequence and N sequence bind.
- The 3' end of N is extended by the action of polymerase, increasing P.
- (The reaction in which P forms a double strand and the reaction in which P dissociates into a single strand are reversible.)
Reaction 3 (Reproducing the life time of N and P)
- Exonuclease decomposes P and N, but not G
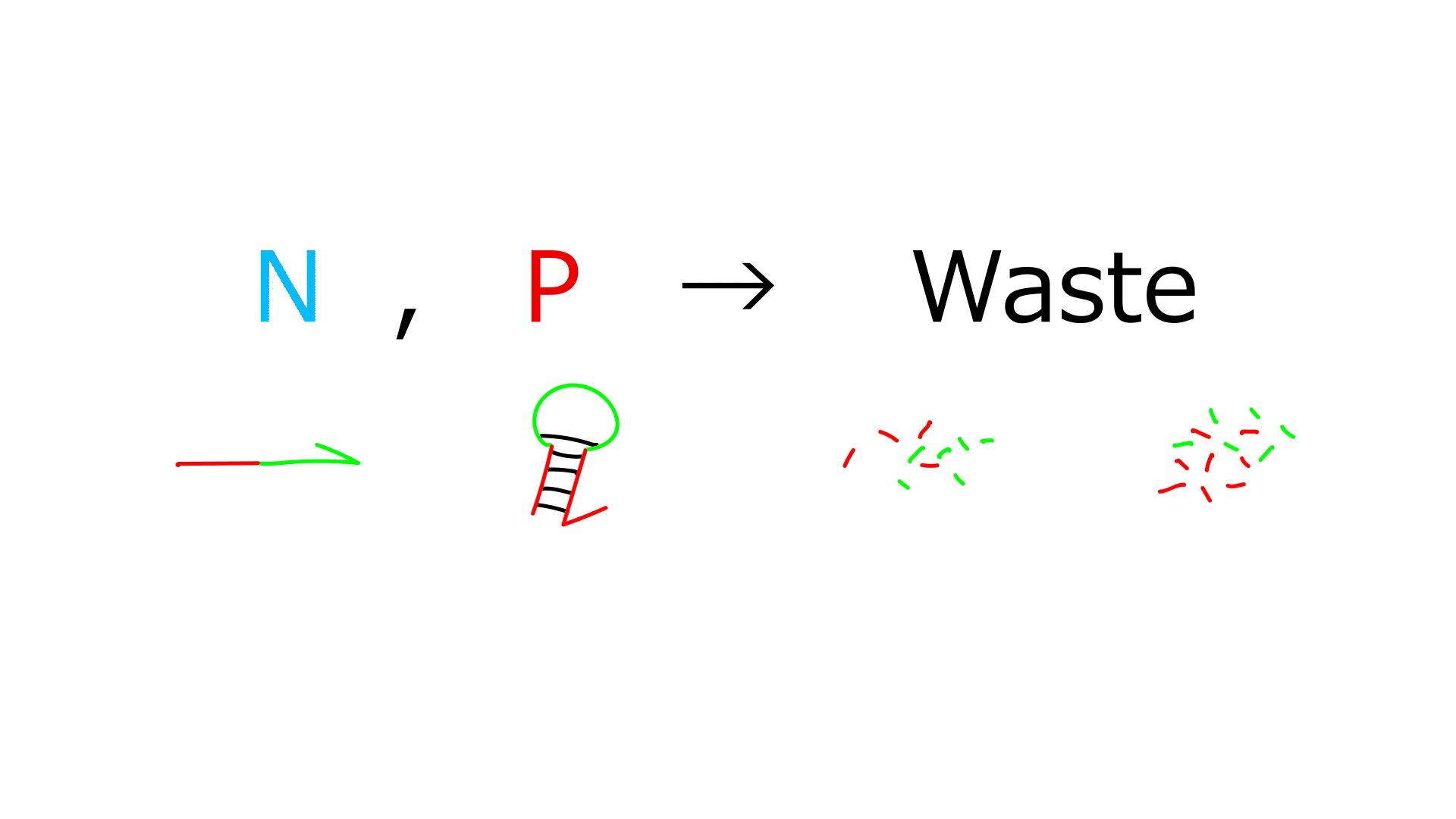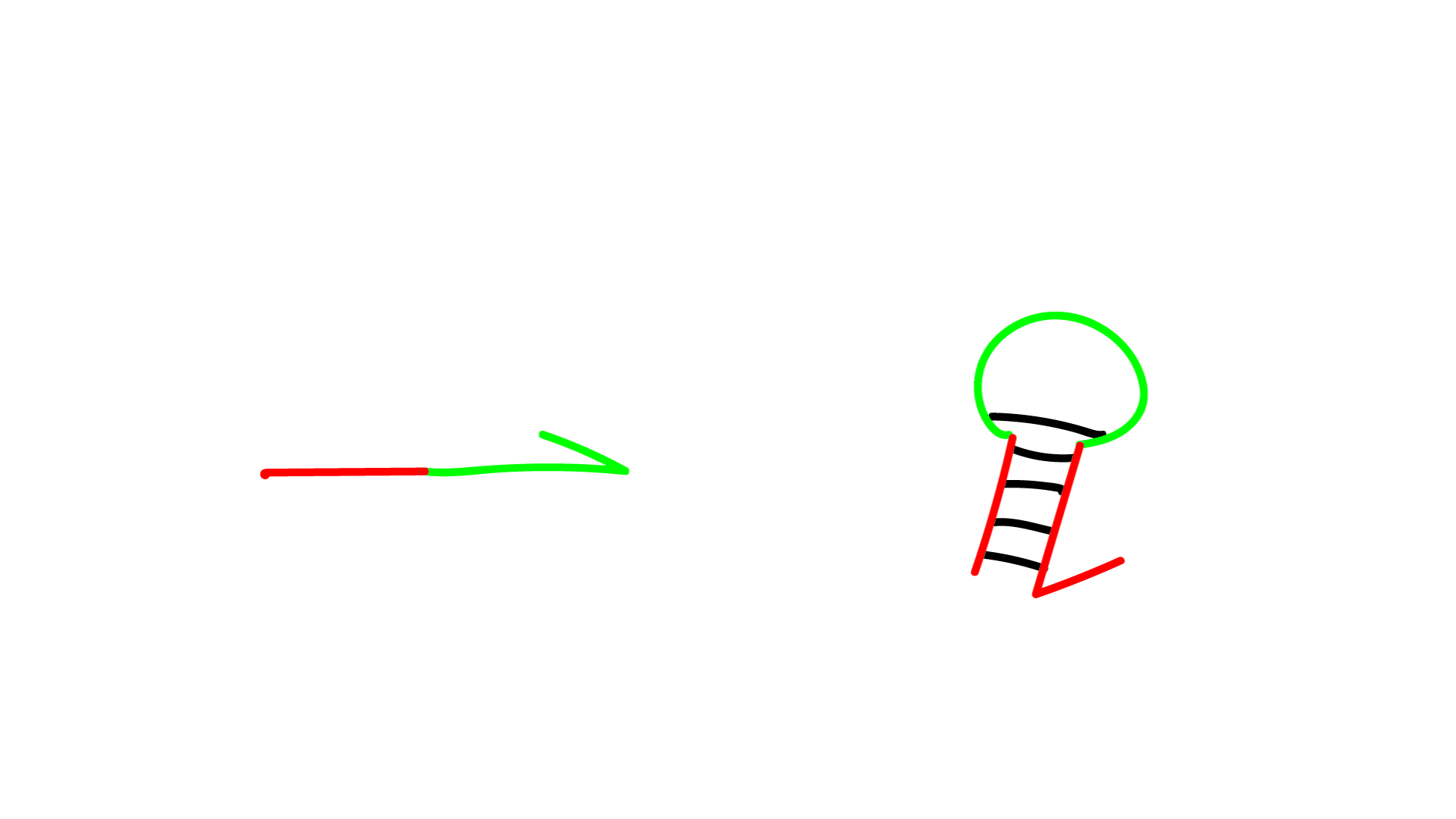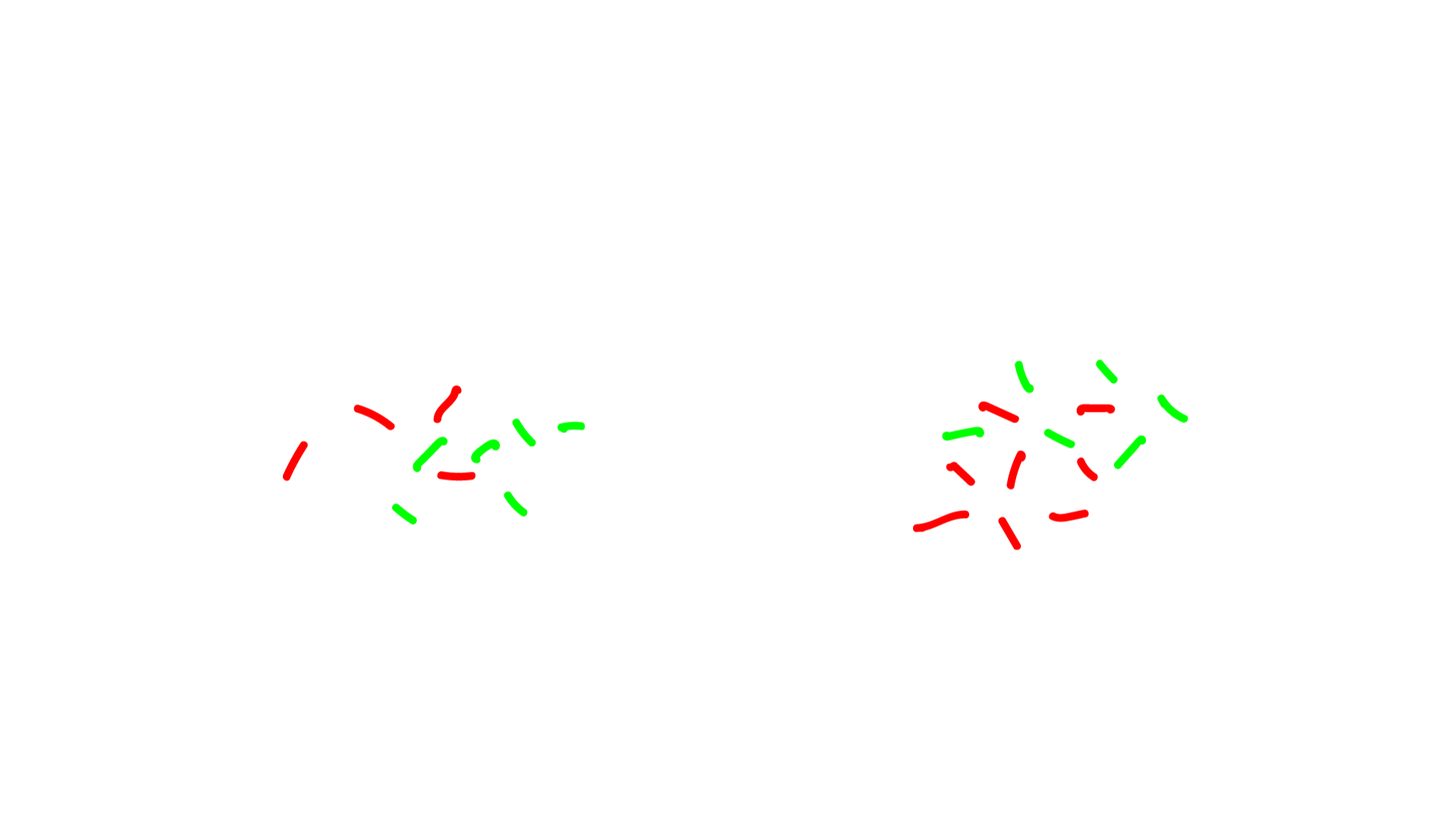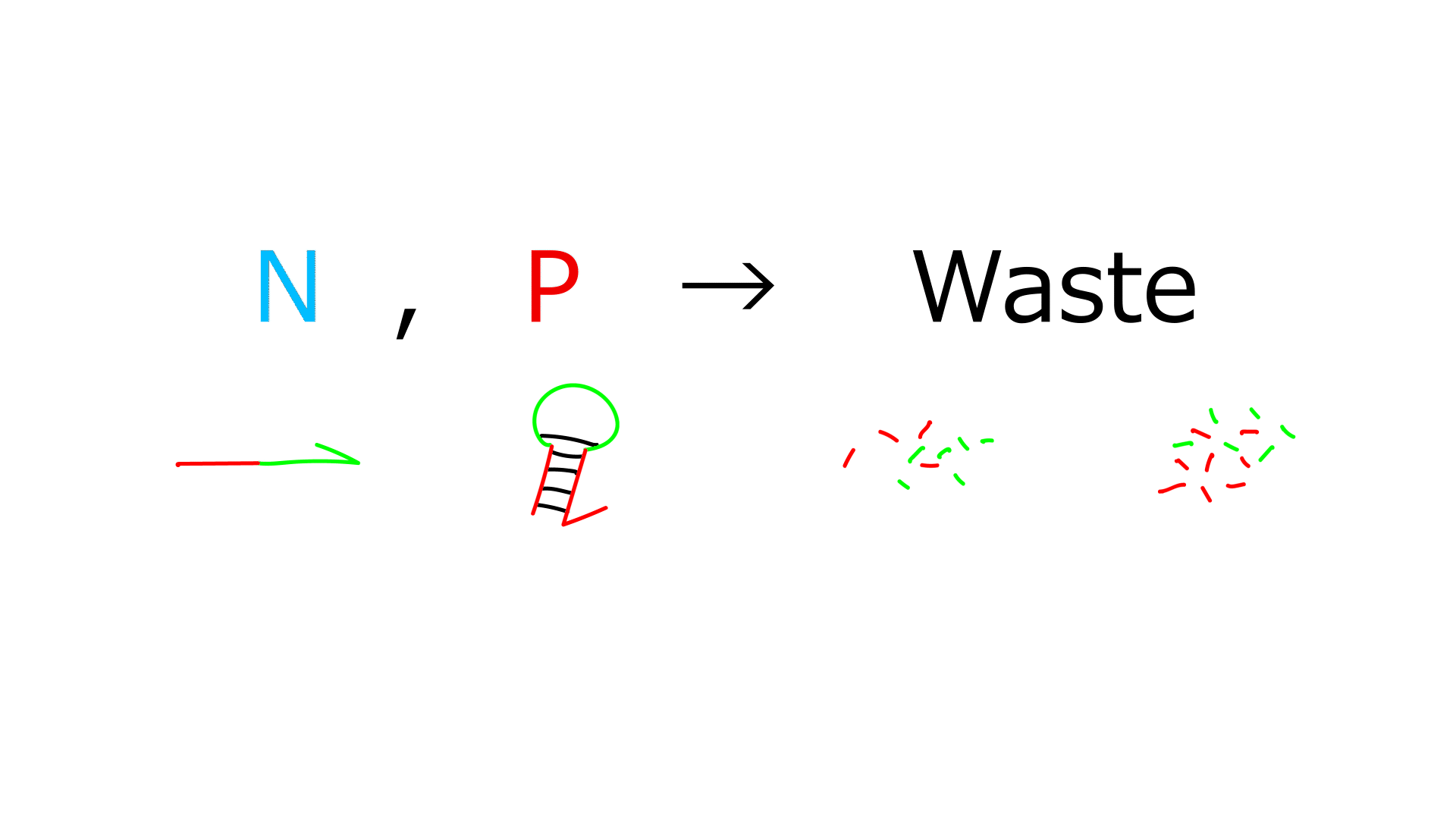
DNA-oscillation Result
The results of combining Reactions 1-3 are shown below.
Amino acid modifications
We utilized guanine modified with an amino group at the 3' terminus to conjugate amino acids. In this process, the amino group of each amino acid was protected with a Boc (tert-butoxycarbonyl) group.
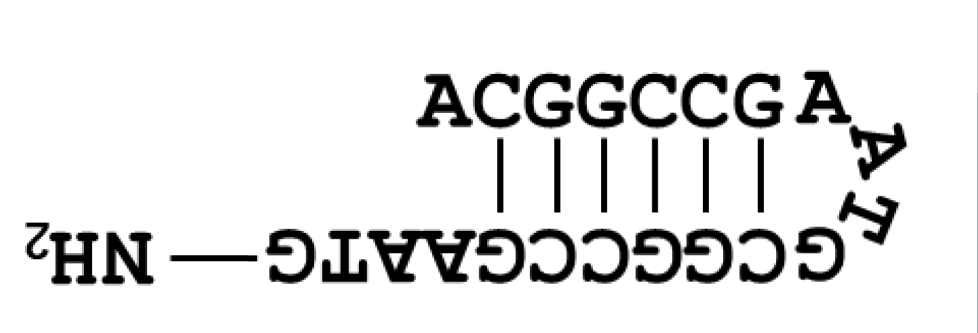
It is anticipated that modification with amino acids will influence the reaction rate between the amino group and guanine, thereby affecting the cycle of the DNA oscillation. Depending on the physicochemical properties of each amino acid, the following effects are expected:
- Positively charged amino acids may enhance DNA binding through electrostatic attraction.
- Negatively charged amino acids may inhibit DNA binding due to Coulombic repulsion.
- Hydrophobic amino acids may influence binding through hydrophobic interactions that attract the hydrophilic DNA.
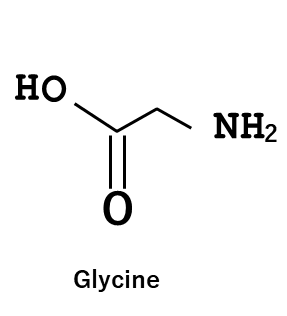
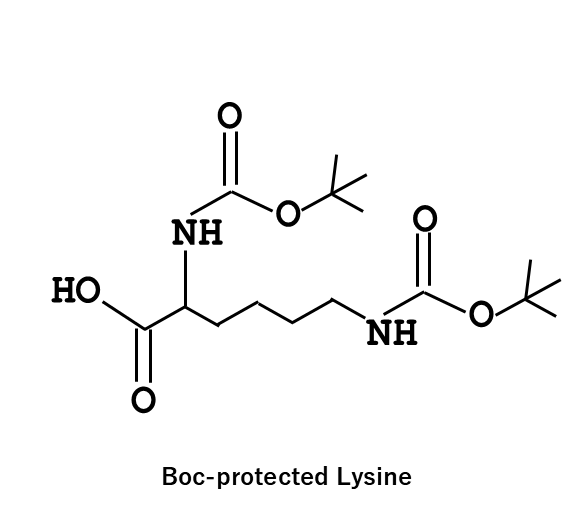
In this study, experiments were conducted using Boc-protected lysine and deprotected glycine. However, it was found that DNA degradation occurred when trifluoroacetic acid (TFA) was used during the Boc deprotection process.
Amino acid modifications result
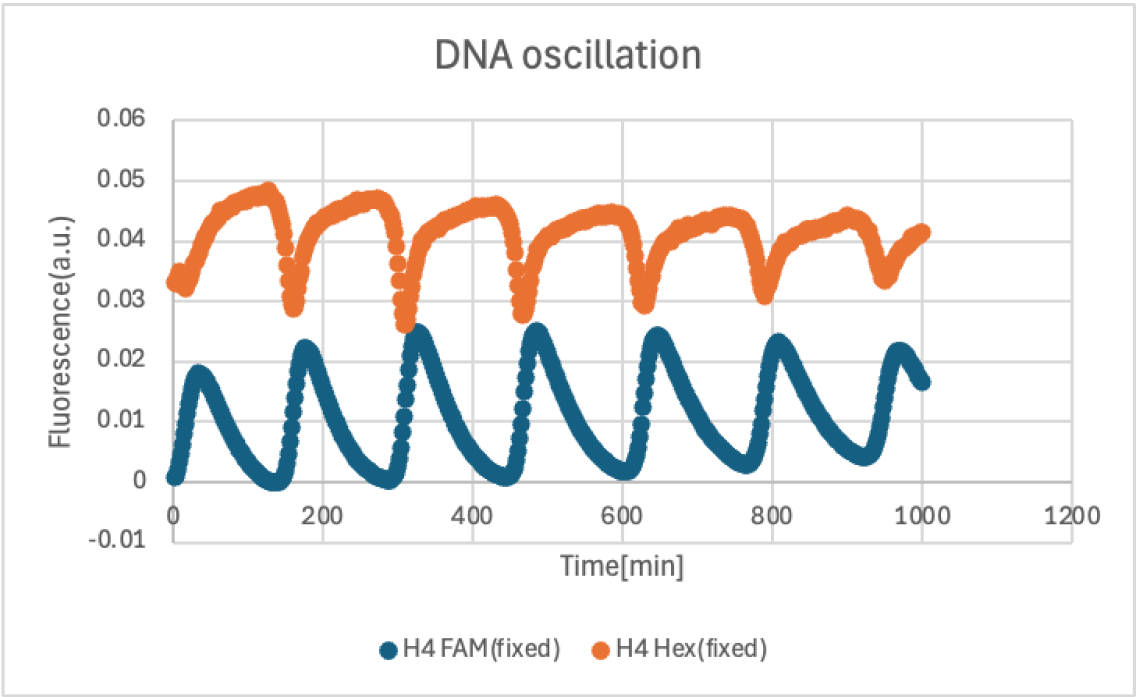
- Oscillatory behavior driven by each template DNA was examined.
- The oscillation generated by the normal G strand, G modified with an amino group, and G modified with Boc-protected lysine were investigated for comparison.
- The reason why the use of the Boc (tert-butoxycarbonyl group)-protected lysine was that the deprotection of Boc by TFA induced the decomposition of the G strand and the deprotection process did not proceed as intended.
- Measurements were performed using real-time PCR at an isothermal temperature of 46 °C for 12 hours.
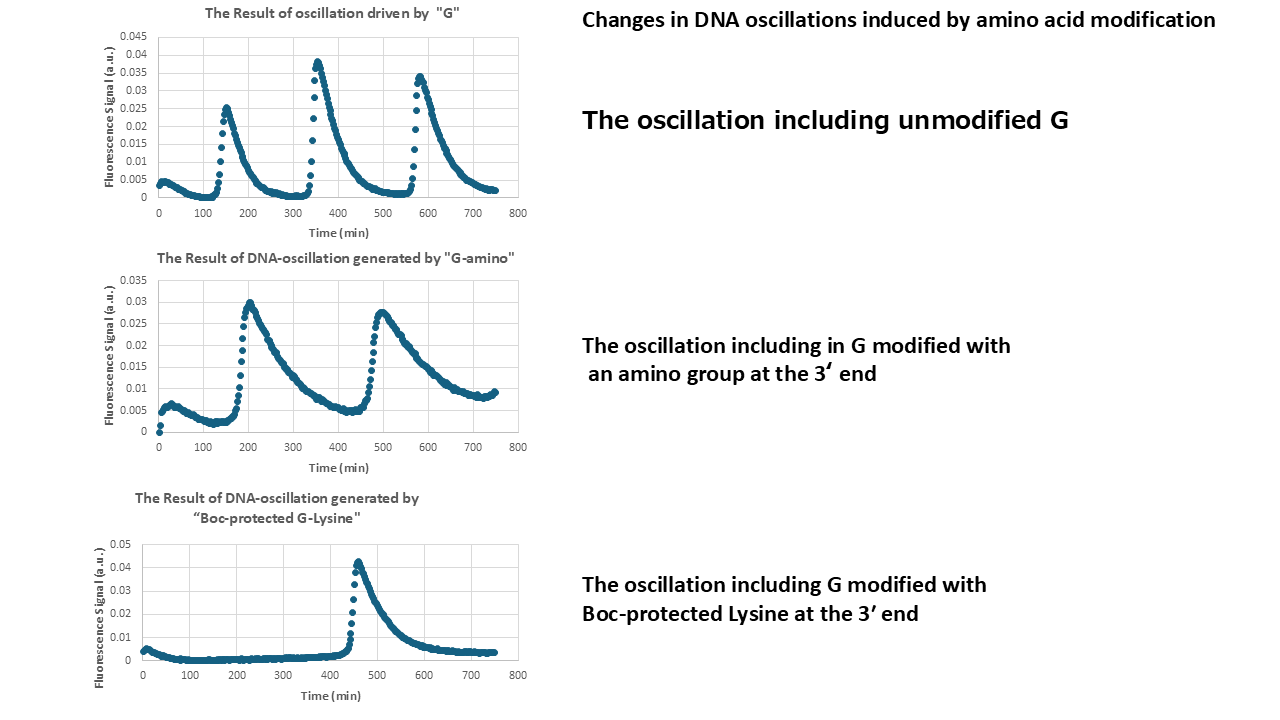
- Oscillatory behavior varied depending on each G strand.
- The oscillation generated by the G strand modified with an amino group exhibited a longer period compared to that of the unmodified G.
- In the case of the G strand modified with Boc-protected lysine, oscillatory behavior was observed only once.
- Here, we discuss the mechanism underlying the differences in oscillatory behavior. G modified with amine would have an additional cationic charge due to the protonation of the amine group. Before performing the experiment, we expected that this additional cation at the 3 prime of G would enhance the hybridization of G and N, leading to acceleration of Reaction 1. Indeed, our simulator also suggested increased association of G and N shortened the period of the oscillation (you can check it, too!). However, the experimental result showed that the period became longer. Therefore, the amine modification to G may decrease the association rate of G and N, which agrees with the simulator results. G modified with Boc-protected lysine showed a single pulse, suggesting that Reaction 1 is strongly inhibited. Because the Boc has three methyl groups, the G has hydrophobic groups in its 3 prime end. Thus, CH-π interaction can occur between Boc and DNA bases. This hydrophobic interaction may alter the secondary structure of G, thereby strongly inhibiting hybridization with N.
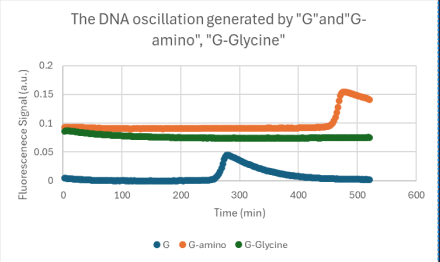
- The results obtained using glycine are shown in the figure above.
- Although distinct pulses could not be observed in comparison with those obtained using Boc-protected lysine, a slight but similar oscillatory behavior was detected.
- These findings indicate that our approach can modulate the characteristics of the oscillation.
For Detailed Experimental Information
Complete protocols, reagent lists, instrument settings, raw/processed data, and extended experimental analyses are available in the supplementary materials.
References
[1] Teruo Fujii, Yannick Rondelez. Predator-Prey Molecular Ecosystems. ACS Nano 2013, 7, 1, 27–34
[2] Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol Syst Biol. 2011 Feb 1;7:466. doi: 10.1038/msb.2010.120. Erratum in: Mol Syst Biol. 2011 Mar 8;7:476.
[3] Aubert-Kato, N., Cazenille, L. Designing Dynamical Molecular Systems with the PEN Toolbox. New Gener. Comput. 38, 341–366 (2020). https://doi.org/10.1007/s00354-020-00089-w
[4] [BIOMOD Cheat Sheet] BIOMOD toranomaki (in Japanese). CBI gakkai (CBI academic conference). 2019;