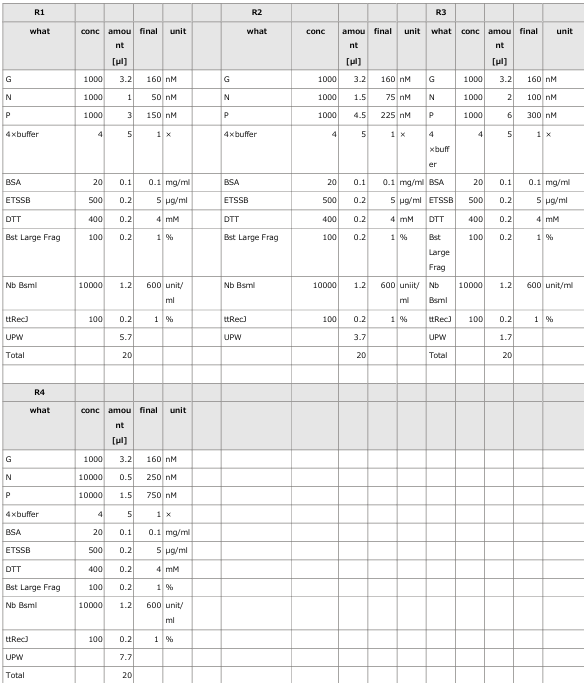
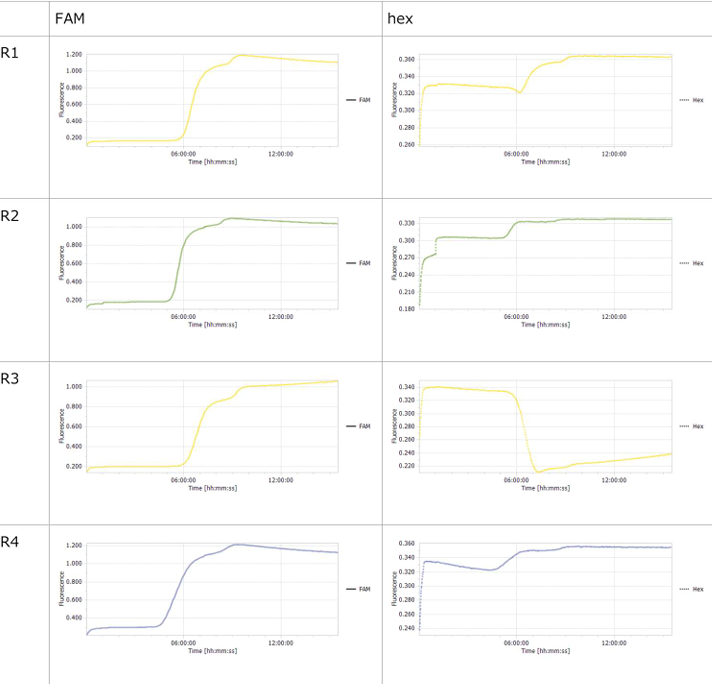
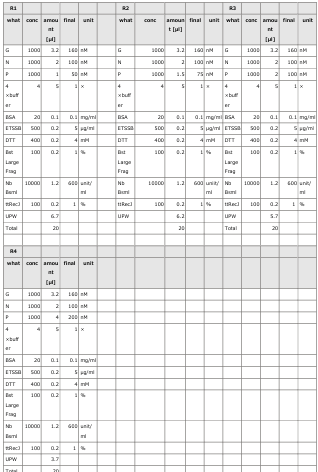
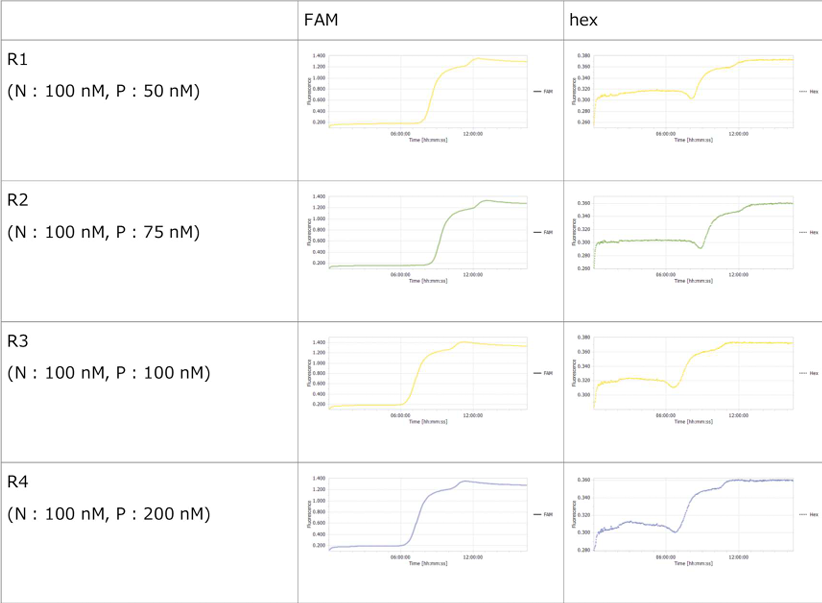
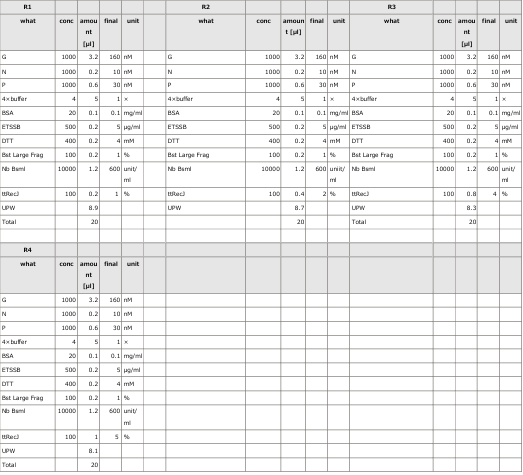
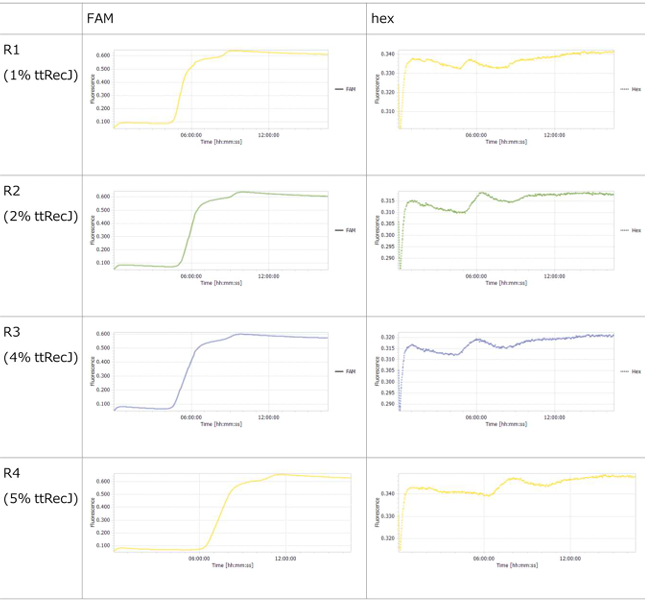
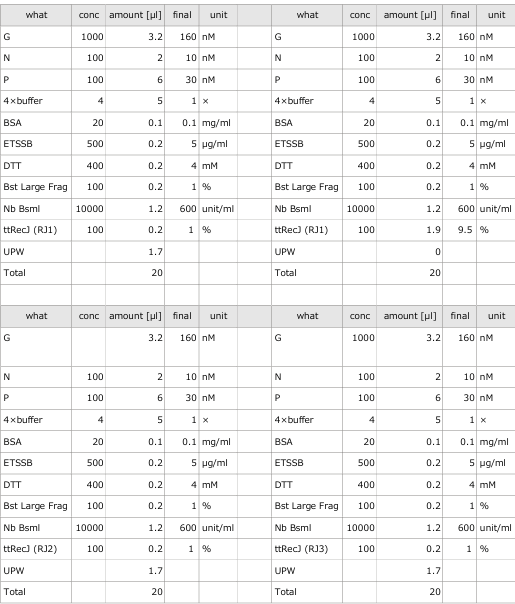
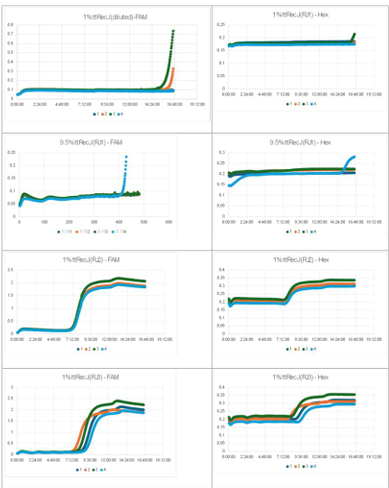
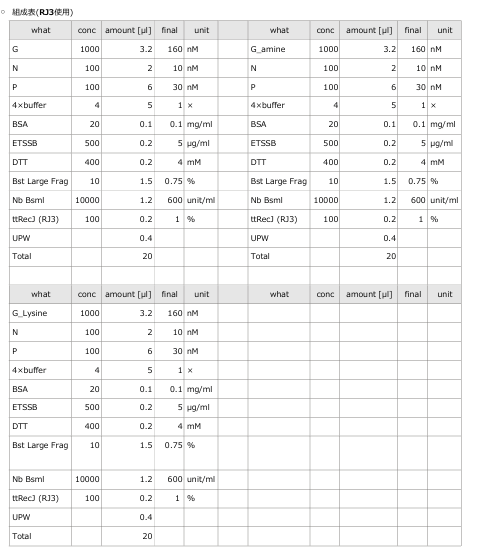
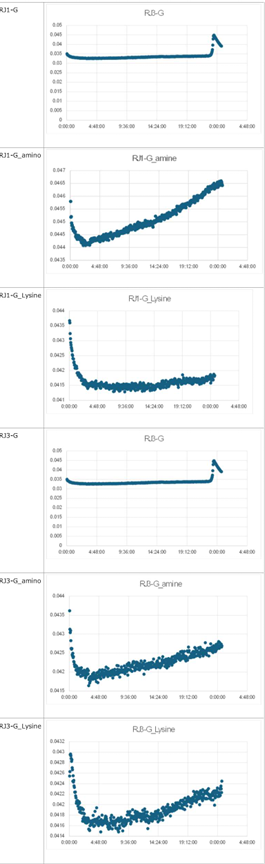
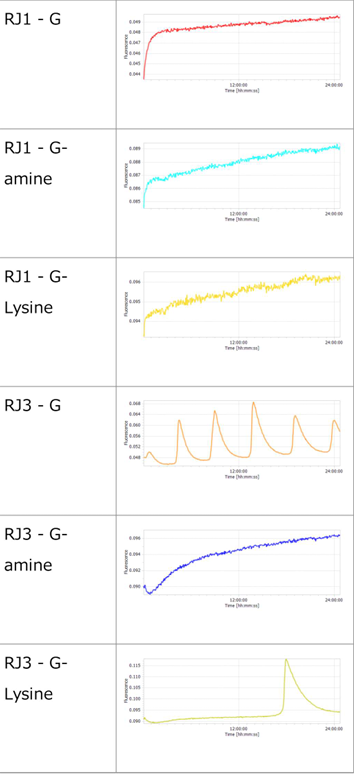
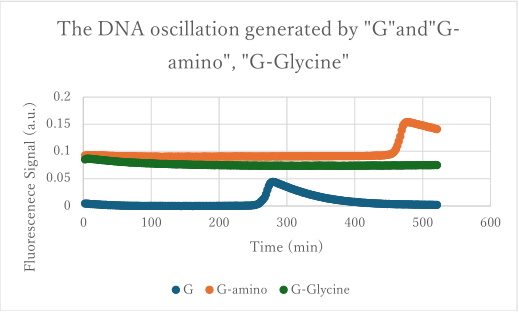
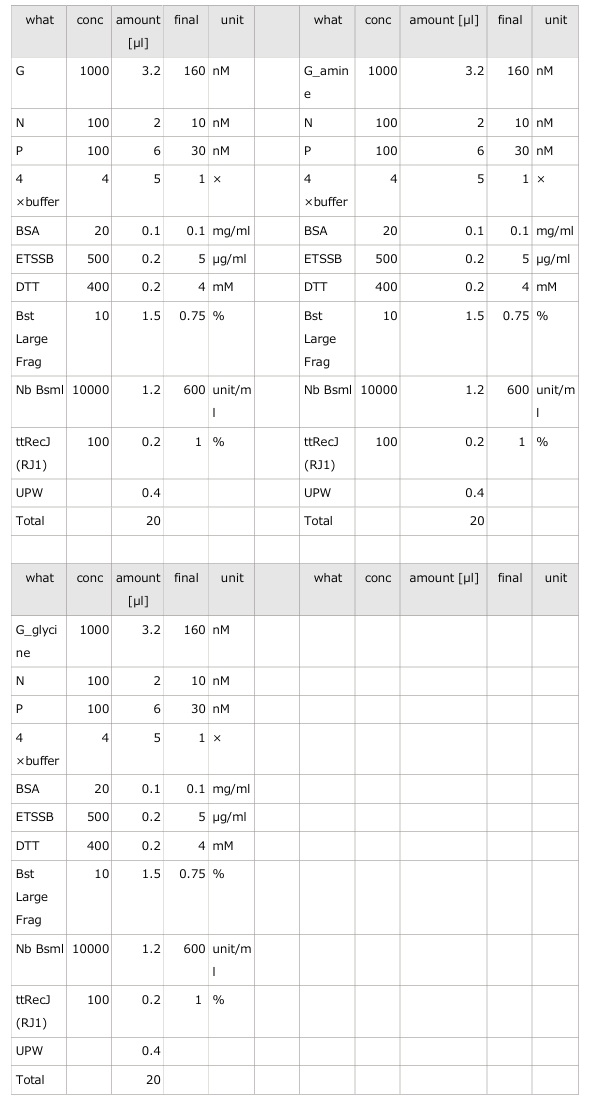
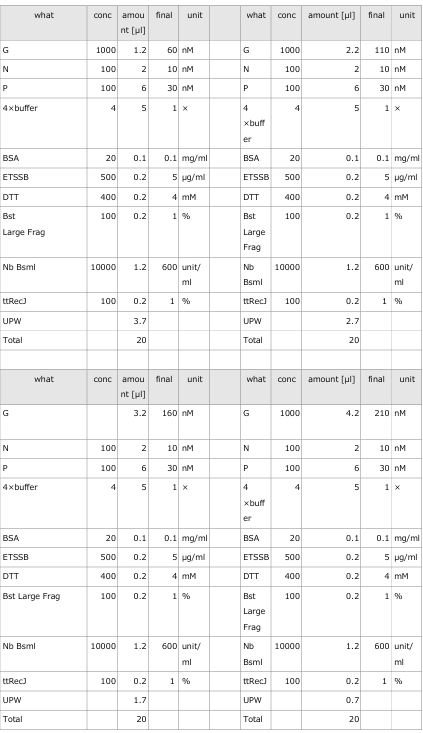
3.1 Experiment 3-1
Objective of the experiment;
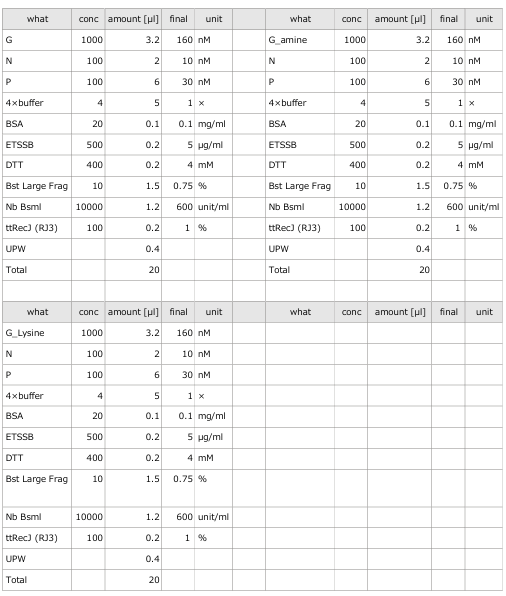
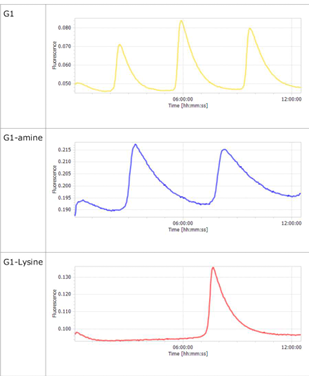
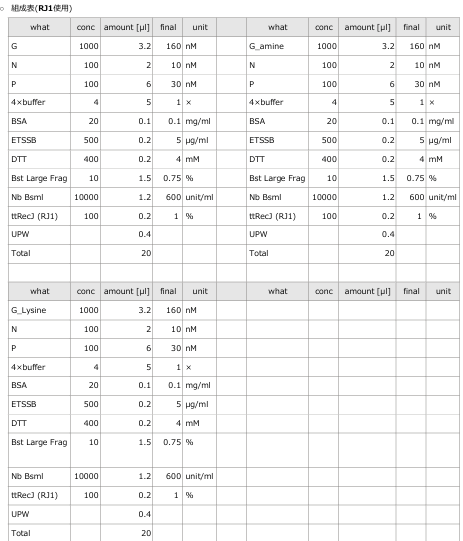
- The oscillations were investigated using the composition parameters described in the previous study [1].
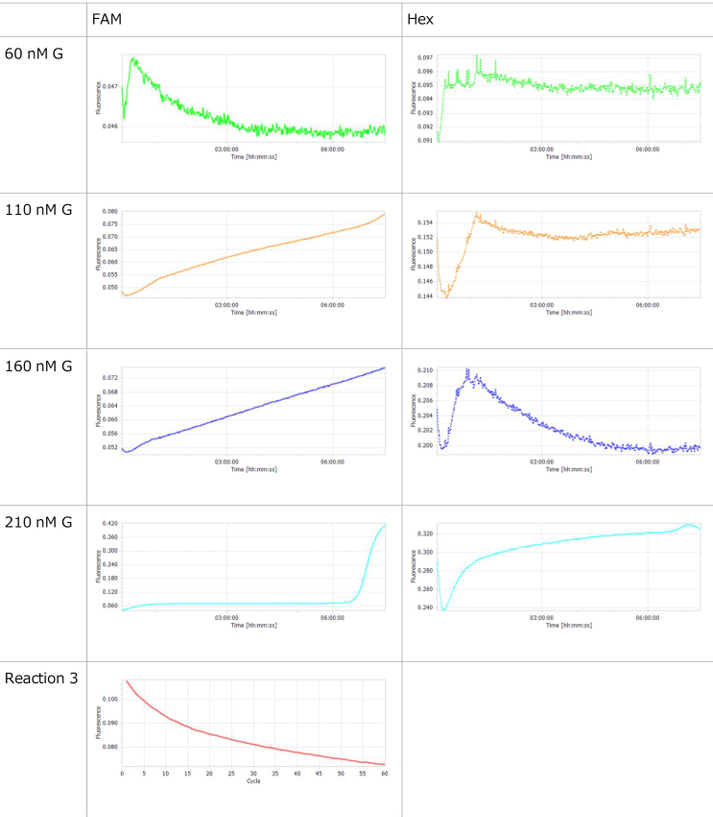
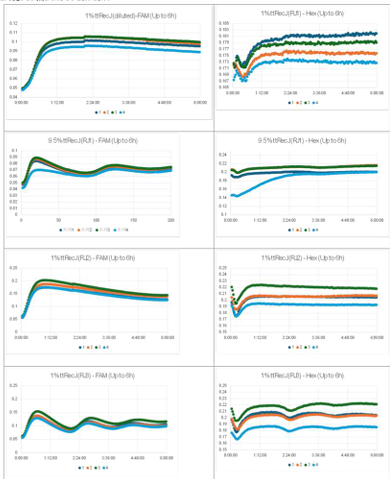
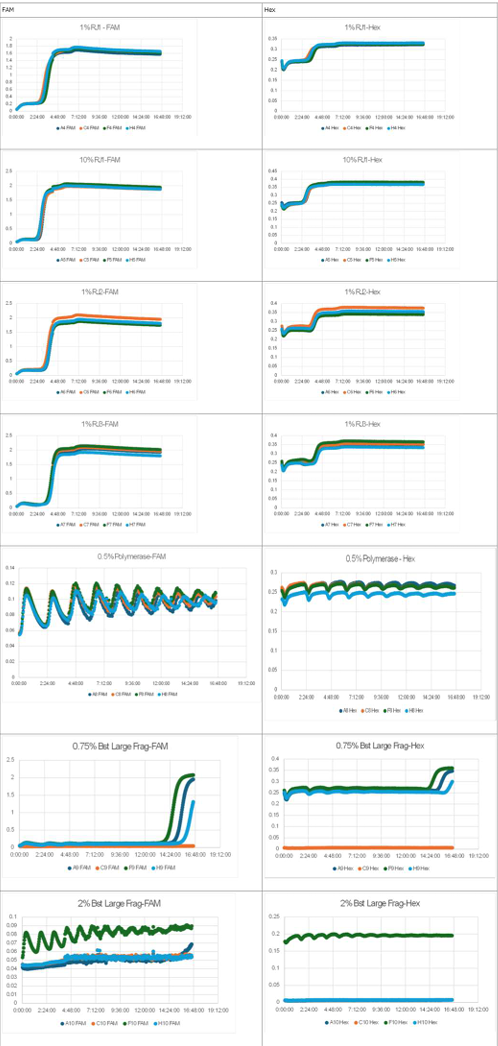
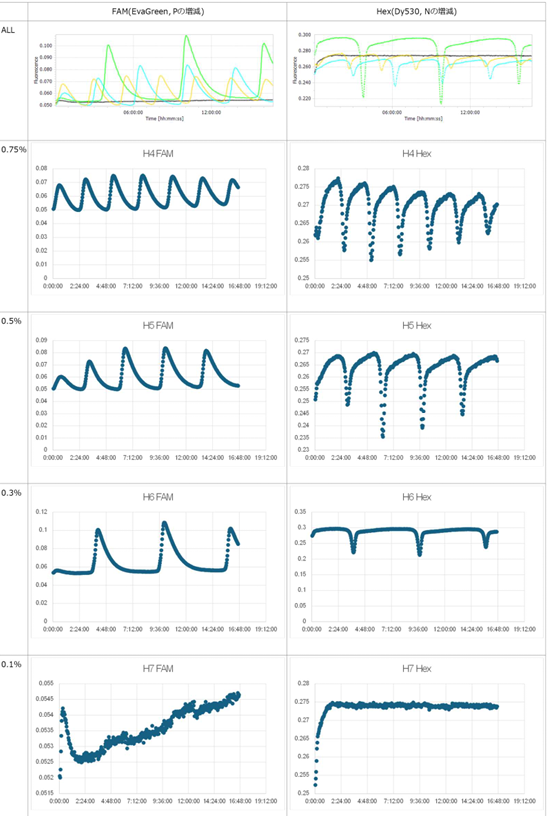
- Horizontal axis: Time; Vertical axis: Fluorescence intensity.
- The graph on the left represents P, while the graph on the right represents N.
- Oscillatory behavior was examined under conditions where the concentration of the template DNA, G, was set to 60 nM, 110 nM, 160 nM, and 210 nM, respectively.
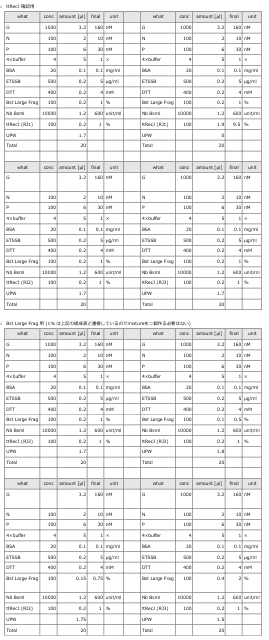
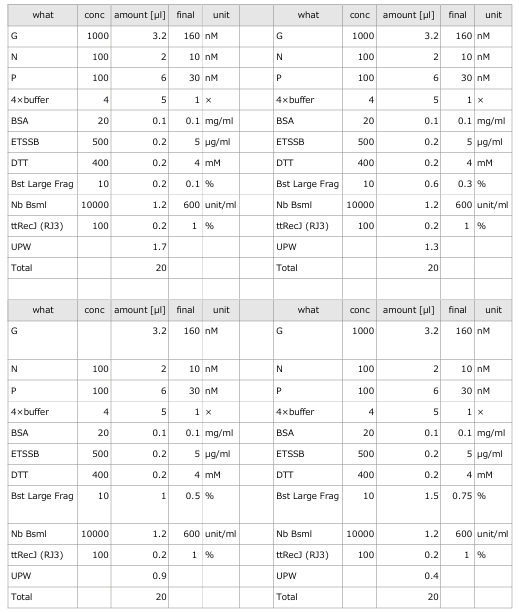
- To evaluate the enzymatic activity of ttRecJ, an independent experiment was performed using Reaction 3 alone.
- Oscillatory behavior was observed immediately after the reaction started; however, the concentration of P subsequently increased. In other words, P did not decrease, while the proportion of N declined.